Software
Check out our software projects at the lab repo and Dr. Zhu’s personal repo.
scikit-bio
scikit-bio is an open-source Python package for bioinformatics researchers and developers. It provides algorithms and data structures for sequence alignments, phylogenetic trees, distance matrices, ordinations and diversity metrics. It powers QIIME 2, Qiita and multiple other bioinformatics tools. Funded by the U.S. Department of Energy (#DE-SC0024320), we are expanding the development of scikit-bio to support efficient multiomic data integration and complex community modeling.
Web of Life (WoL)
The WoL project aims at building a reference phylogeny which accurately defines the evolutionary relationships among all microbes. In Phase I of the project, we built a phylogeny of 10,575 genomes using 381 marker genes, making this the single largest dataset upon which de novo phylogenetic trees had been built, yet the bioinformatic approaches we adopted or invented are significantly more robust than previous works. It means to serve as a reference for researchers to explore the evolution and diversity of microbes, and to improve the study of microbial communities.
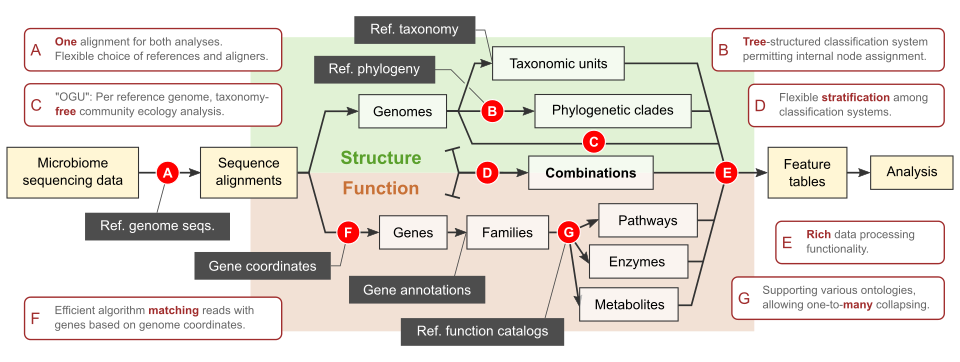
Woltka
Woltka is a bioinformatics package for shotgun metagenomic data analysis. It highlights: 1) fine-grain community ecology featuring individual reference genomes; 2) tree-based, rank-free classification to maximize resolution and flexibility; 3) combined taxonomic & functional analysis through one alignment to ensure consistency and accuracy. It takes full advantage of, but not limited by, the WoL reference phylogeny. It comes with an interface for the QIIME 2 package, and has been integrated into the Qiita web server.
BinaRena
Binarena (“bin arena”) is an interactive visualizer and operator of metagenomic contigs to facilitate discovery of biological patterns and recovery of MAGs. It is dedicated to human-guided research in order to complement algorithmic workflows. It lets the user conveniently observe various characteristics of large metagenomic datasets, efficiently manipulate contig-bin assignments, and calculate bin quality metrics in real time. BinaRena is an installation-free, client-end web application. Here is a live demo.
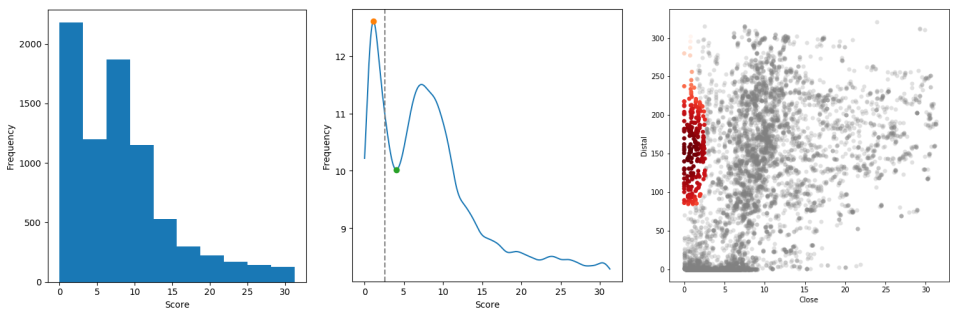
HGTector2
HGTector is a pipeline for genome-wide detection of putative horizontal gene transfer (HGT) events based on sequence homology search hit distribution statistics. HGTector2 is a completely re-engineered software tool, featuring a fully automated analytical pipeline with smart determination of parameters which requires minimum human involvement, a re-designed command-line interface which facilitates standardized scientific computing, and a high-quality Python 3 codebase.
QIIME 2
QIIME 2 is an integrated software package for microbiome data analysis. It provides a complete and flexible solution from raw sequencing data to publication-grade tables and figures. It highlights transparent and reproducible science. It has been the most widely-used bioinformatics tool in the field of microbiomics. We actively contribute to the QIIME 2 ecosystem.
Utilities
A collection of single-file scripts written by Dr. Zhu. There might be useful things in it…